Bacteriophage T4 Atomistic Model
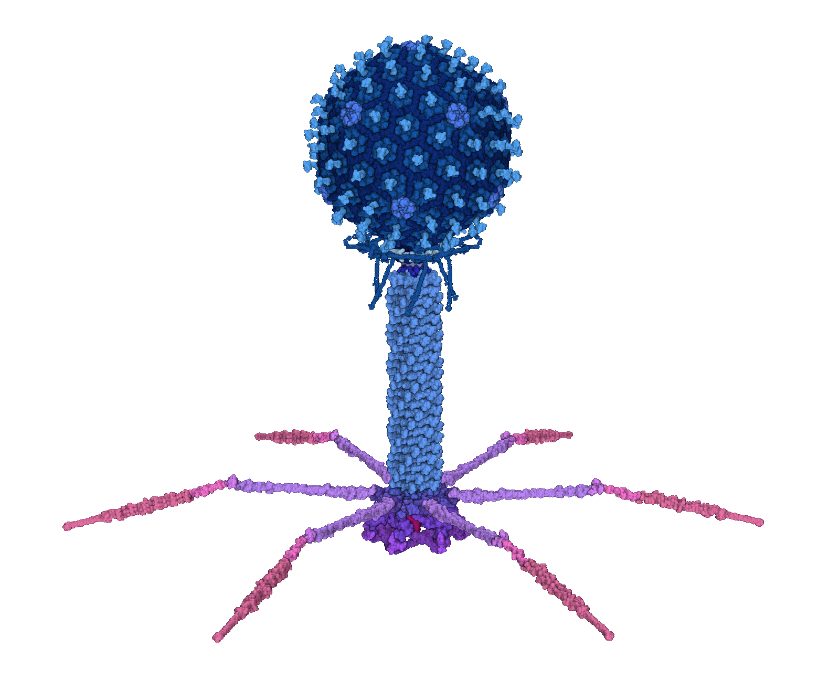
Bacteriophage T4 isometric type
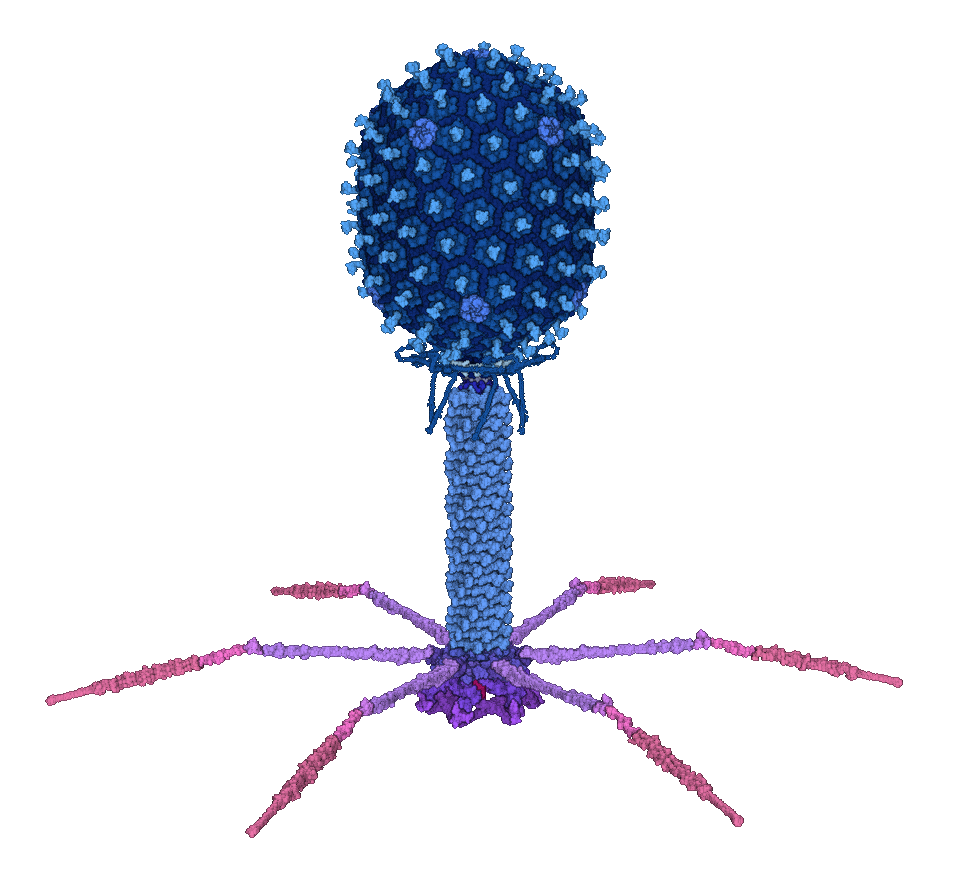
Bacteriophage T4 wild type
Authors
Aeliya Syed, John Winfer, Leon Thistle, Paul Ekers, Ngan Nguyen, Ondrej Strnad, David Goodsell, Ivan Viola, Deng Luo.Description
This is a WISE [1] internship project aimed at building the atomistic model for bacteriophage T4. The most up-to-date structural data and knowledge are first collected and then fed into the in-house modeling tool called MesoCraft. Most proteins' structures are collected from the PDB database. When this is not available, AlphaFold2 and SWISS-MODEL are the main tools used to perform the prediction of the protein's structure from its amino acid sequence. Please refer to the "YouTube - Bacteriophage T4 Atomistic Model Structural Biology Background" link below for more information.
The above banner image shows the isometric mutant type T4, whose head capsid is a perfect icosahedron with the triangulation number T=13. Its model can be downloaded in the "Isometric_mutant_type_T4_v3.zip" link below. The model is released in MesoCraft .model format. A .cif format will be released later. The wild type T4 is being constructed, with T=13 forming the top and bottom cap and T=20 forming the middle section.
The model is subject to update when new insights are found. Please address correspondence to Deng Luo.
1. WISE internship program: https://tks.kaust.edu.sa/beyond-the-classroom/secondary/wise
Downloads & Resources
- Isometric_mutant_type_T4_v3.zip
- YouTube - Bacteriophage T4 Atomistic Model Structural Biology Background
- Atomistic model of an Escherichia virus T4 bacteriophage
- Video - Bacteriophage T4 Atomistic Model Structure
- WISE Presentation Slides: Nanovis 3D Modelling Internship
How to cite & Licence

@misc{NanovisG51:online,
author = {John Winfer, Aeliya Syed, Paul Ekers, Leon Thistle, Ngan Nguyen, Ondrej Strnad, David Goodsell, Ivan Viola, Deng Luo},
title = {T4 model},
howpublished = {\url{https://www.nanovis.org/T4-model.html}},
month = {},
year = {},
note = {(Accessed on 09/07/2021)}
}