Towards Differentiable Electron Microscopy Simulation: Methods and Applications
Authors
Ngan Nguyen, Feng Liang, Dominik Engel, Ciril Bohak, Ondřej Strnad, Timo Ropinski, Ivan ViolaDescription
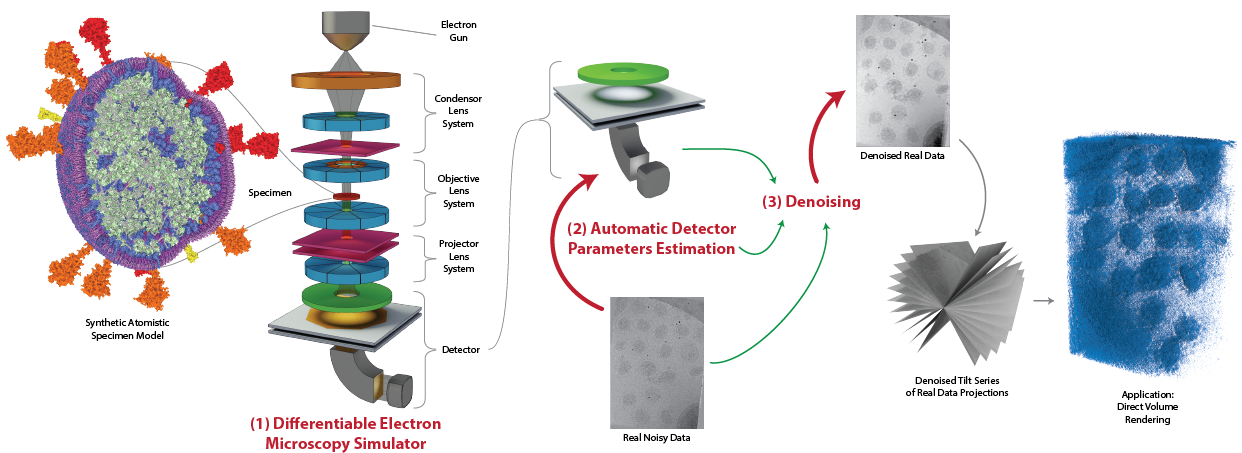
We propose a new microscopy simulation system that can depict atomistic models in a micrograph visual style, similar to physical electron microscopy imaging results. This system is scalable, able to represent the simulation of electron microscopy of twenty complex viral particles, and synthesizes the image faster than previous approaches. Additionally, the simulator is differentiable in the deterministic and stochastic stages that form signal and noise representations in the micrograph. This allows for solving inverse problems by means of optimization and thus allows for the generation of microscopy simulations using parameter settings estimated from real data. We demonstrate this capability through two applications: (1) estimating the parameters of the modulation transfer function defining the detector properties of the simulated and real micrographs and (2) denoising the real data based on parameters optimized from the simulated examples using gradient descent. While current simulators do not support any parameter estimation due to their forward-only design, we show that the results obtained using estimated parameters are very similar to real micrographs. Additionally, we evaluate the denoising capabilities of our approach and show that the results are competitive with state-of-the-art methods. Denoised micrographs exhibit less noise in the tomographic reconstruction of tilt-series, facilitating visualization of microscopy tomography using direct volume rendering by reducing the visual dominance of noise.
Sources